Saturday, December 31, 2022
Keeping Up With The AI Competition
Thursday, December 29, 2022
Mehmed v. Vlad
Tuesday, December 27, 2022
Genetics Favor Domestication Of Cats In The Near East
Cat domestication likely initiated as a symbiotic relationship between wildcats (Felis silvestris subspecies) and the peoples of developing agrarian societies in the Fertile Crescent. As humans transitioned from hunter-gatherers to farmers ~12,000 years ago, bold wildcats likely capitalized on increased prey density (i.e., rodents). Humans benefited from the cats’ predation on these vermin. To refine the site(s) of cat domestication, over 1000 random-bred cats of primarily Eurasian descent were genotyped for single-nucleotide variants and short tandem repeats. The overall cat population structure suggested a single worldwide population with significant isolation by the distance of peripheral subpopulations. The cat population heterozygosity decreased as genetic distance from the proposed cat progenitor’s (F.s. lybica) natural habitat increased. Domestic cat origins are focused in the eastern Mediterranean Basin, spreading to nearby islands, and southernly via the Levantine coast into the Nile Valley. Cat population diversity supports the migration patterns of humans and other symbiotic species
Friday, December 23, 2022
The Aztec Calendar Worked
Without clocks or modern tools, ancient Mexicans watched the sun to maintain a farming calendar that precisely tracked seasons and even adjusted for leap years.
Thursday, December 22, 2022
Another Global Constraint On BSM Physics
The Standard Model cannot explain the dominance of matter over anti-matter in our universe. This imbalance indicates undiscovered physics that violates combined CP symmetry.
Many extensions to the Standard Model seek to explain the imbalance by predicting the existence of new particles. Vacuum fluctuations of the fields associated with these new particles can interact with known particles and make small modifications to their properties; for example, particles which violate CP symmetry will induce an electric dipole moment of the electron (eEDM). The size of the induced eEDM is dependent on the masses of the new particles and their coupling to the Standard Model. To date, no eEDM has been detected, but increasingly precise measurements probe new physics with higher masses and weaker couplings.
Here we present the most precise measurement yet of the eEDM using electrons confined inside molecular ions, subjected to a huge intra-molecular electric field, and evolving coherently for up to 3 seconds. Our result is consistent with zero and improves on the previous best upper bound by a factor ∼2.4. Our sensitivity to 10^−19 eV shifts in molecular ions provides constraints on broad classes of new physics above 10^13 eV, well beyond the direct reach of the LHC or any other near- or medium-term particle collider.
Native American Solstice Ceremonies
The winter solstice is the day of the year when the Northern Hemisphere has the fewest hours of sunlight and the Southern Hemisphere has the most. For indigenous peoples, it has been a time to honor their ancient sun deity. They passed their knowledge down to successive generations through complex stories and ritual practices.
An Insight Into QCD Math Has The Potential To Rock QCD Phenomenology
We show that the topological charge of nonabelian gauge theory is unphysical by using the fact that it always involves the unphysical gauge field component proportional to the gradient of the gauge function. The removal of Gribov copies, which may break the Becchi-Rouet-Stora-Tyutin symmetry, is irrelevant thanks to the perturbative one-loop finiteness of the chiral anomaly. The unobservability of the topological charge immediately leads to the resolution of the Strong CP problem. We also present important consequences such as the physical relevance of axial U(1) symmetry, the θ-independence of vacuum energy, the unphysicalness of topological instantons, and the impossibilities of realizing the sphaleron induced baryogenesis as well as the chiral magnetic effect. The unphysical vacuum angle and the axial U(1) symmetry also imply that the CP phase of the Cabibbo-Kobayashi-Maskawa matrix is the sole source of CP violation of the standard model.
Wednesday, December 21, 2022
The Triumphant Standard Model
Tuesday, December 20, 2022
The Sumerian Roots Of the Menorah
The Old European Culture blog in yet another thought provoking post muses on possible Sumerian roots of the Menorah in Jewish culture.
Lepton Universality Violations Have Disappeared With Improved LHC Data
The first simultaneous test of muon-electron universality using B+→K+ℓ+ℓ− and B0→K∗0ℓ+ℓ− decays is performed, in two ranges of the dilepton invariant-mass squared, q2. The analysis uses beauty mesons produced in proton-proton collisions collected with the LHCb detector between 2011 and 2018, corresponding to an integrated luminosity of 9 fb−1. Each of the four lepton universality measurements reported is either the first in the given q2 interval or supersedes previous LHCb measurements. The results are compatible with the predictions of the Standard Model.
A simultaneous analysis of the B+→K+ℓ+ℓ− and B0→K∗0ℓ+ℓ− decays is performed to test muon-electron universality in two ranges of the square of the dilepton invariant mass, q2. The measurement uses a sample of beauty meson decays produced in proton-proton collisions collected with the LHCb detector between 2011 and 2018, corresponding to an integrated luminosity of 9 fb−1. A sequence of multivariate selections and strict particle identification requirements produce a higher signal purity and a better statistical sensitivity per unit luminosity than previous LHCb lepton universality tests using the same decay modes. Residual backgrounds due to misidentified hadronic decays are studied using data and included in the fit model. Each of the four lepton universality measurements reported is either the first in the given q2 interval or supersedes previous LHCb measurements. The results are compatible with the predictions of the Standard Model.
Saturday, December 17, 2022
LHC To Announce Its Lepton Universality Violation Results On Tuesday
Experimental results tending to show lepton universality violations (i.e. a different probability for decays to tau leptons, muons, and electron-positron pairs respectively, mass-energy conservation permitting) are the most notable experimental anomalies from Standard Model predictions outstanding right now in high energy physics.
But the statistical significance is merely a tension that may fade with a major new data point like the one to be announced on Tuesday, and there isn't a good explanation for why it isn't seen in other phenomena that should involve the same intermediate W boson decay driven processes.
Measurements of 𝑅(𝐾) and 𝑅(𝐾∗) with the full LHCb Run 1 and 2 databy Renato Quagliani (EPFL - Ecole Polytechnique Federale Lausanne (CH))In this seminar we present the first simultaneous test of muon-electron universality in 𝐵+→𝐾+ℓ+ℓ− and 𝐵0→𝐾∗0ℓ+ℓ− decays, known as 𝑅(𝐾) and 𝑅(𝐾∗), in two regions of di-lepton invariant mass squared.The analysis operates at a higher signal purity compared with previous analyses and implements a data-driven treatment of residual hadronic backgrounds. The analysis uses the full LHCb Run 1 and 2 data recorded in 2011-2012 and 2015-2018, corresponding to an integrated luminosity of 9 fb−1. This analysis is the most sensitive lepton universality test in rare b-decays and the results obtained supersede the previous LHCb measurements of 𝑅(𝐾) and 𝑅(𝐾∗0).
Tuesday, December 13, 2022
Old Water
Water is crucial for the emergence and evolution of life on Earth. Recent studies of the water content in early forming planetary systems similar to our own show that water is an abundant and ubiquitous molecule, initially synthesized on the surfaces of tiny interstellar dust grains by the hydrogenation of frozen oxygen. Water then enters a cycle of sublimation/freezing throughout the successive phases of planetary system formation, namely, hot corinos and protoplanetary disks, eventually to be incorporated into planets, asteroids, and comets. The amount of heavy water measured on Earth and in early forming planetary systems suggests that a substantial fraction of terrestrial water was inherited from the very first phases of the Solar System formation and is 4.5 billion years old.
Friday, December 9, 2022
New mtDNA Clade Discovered
I missed this paper when it came out, but found it now that it has won an editor's choice award, as one of its authors explained at their blog.
As background, the Sandawe people in whom the new clade is found, are East African desert/savanna hunter-gatherers who speak a click language, while the Mbuti people, in whom the sister mtDNA clade L5 is found, are African pygmies found in the Congo jungle (their ancestral language was lost, mostly to Bantu languages, before it was ever attested).
Archaeological and genomic evidence suggest that modern Homo sapiens have roamed the planet for some 300–500 thousand years. In contrast, global human mitochondrial (mtDNA) diversity coalesces to one African female ancestor (“Mitochondrial Eve”) some 145 thousand years ago, owing to the ¼ gene pool size of our matrilineally inherited haploid genome. Therefore, most of human prehistory was spent in Africa where early ancestors of Southern African Khoisan and Central African rainforest hunter-gatherers (RFHGs) segregated into smaller groups. Their subdivisions followed climatic oscillations, new modes of subsistence, local adaptations, and cultural-linguistic differences, all prior to their exodus out of Africa. Seven African mtDNA haplogroups (L0–L6) traditionally captured this ancient structure—these L haplogroups have formed the backbone of the mtDNA tree for nearly two decades.
Here we describe L7, an eighth haplogroup that we estimate to be ~ 100 thousand years old and which has been previously misclassified in the literature. In addition, L7 has a phylogenetic sublineage L7a*, the oldest singleton branch in the human mtDNA tree (~ 80 thousand years). We found that L7 and its sister group L5 are both low-frequency relics centered around East Africa, but in different populations (L7: Sandawe; L5: Mbuti). Although three small subclades of African foragers hint at the population origins of L5'7, the majority of subclades are divided into Afro-Asiatic and eastern Bantu groups, indicative of more recent admixture. A regular re-estimation of the entire mtDNA haplotype tree is needed to ensure correct cladistic placement of new samples in the future.
Thursday, December 8, 2022
The Latest CKM Matrix Fit
The CKM matrix, which can be reduced to four parameters, quantifies the likelihood of quarks transforming into different kinds of quarks in W boson mediated weak force interactions. The latest global fit of those Standard Model parameters are as follows:
A New Branch Of The Tree Of Life
The tree of life is a useful diagram for understanding the relationships between different forms of life, present and extinct. The trunks are made up of three broad groups called domains – Bacteria, Archaea and Eukaryota – which then branch into kingdoms such as animals and fungi. From there the branches become more and more specific until you reach individual species.The new discovery adds quite a major bough to the tree – Provora. These lifeforms make up a category informally called a “supergroup,” which sits below domains and can contain multiple kingdoms.“This is an ancient branch of the tree of life that is roughly as diverse as the animal and fungi kingdoms combined, and no one knew it was there,” said Dr. Patrick Keeling, senior author of the study.Members of the Provora supergroup are tiny organisms that the team describes as the “lions of the microbial world.” That’s because they prey upon other microbes, and within their ecosystem they’re relatively rare. The supergroup is further divided into two clades – the “nibblerids,” which use tooth-like structures to nibble chunks off their prey, and the “nebulids,” which engulf their prey whole.The team discovered this new kind of life in samples taken from around the world, including the coral reefs in Curaçao, sediment from the Black and Red seas and water from the Pacific and Arctic oceans. . . .“In the taxonomy of living organisms, we often use the gene ‘18S rRNA’ to describe genetic difference,” said Dr. Denis Tikhonenkov, first author of the study. “For example, humans differ from guinea pigs in this gene by only six nucleotides. We were surprised to find that these predatory microbes differ by 170 to 180 nucleotides in the 18S rRNA gene from every other living thing on Earth. It became clear that we had discovered something completely new and amazing.”
Molecular phylogenetics of microbial eukaryotes has reshaped the tree of life by establishing broad taxonomic divisions, termed supergroups, that supersede the traditional kingdoms of animals, fungi and plants, and encompass a much greater breadth of eukaryotic diversity. The vast majority of newly discovered species fall into a small number of known supergroups.
Recently, however, a handful of species with no clear relationship to other supergroups have been described, raising questions about the nature and degree of undiscovered diversity, and exposing the limitations of strictly molecular-based exploration.
Here we report ten previously undescribed strains of microbial predators isolated through culture that collectively form a diverse new supergroup of eukaryotes, termed Provora. The Provora supergroup is genetically, morphologically and behaviourally distinct from other eukaryotes, and comprises two divergent clades of predators—Nebulidia and Nibbleridia—that are superficially similar to each other, but differ fundamentally in ultrastructure, behaviour and gene content.
These predators are globally distributed in marine and freshwater environments, but are numerically rare and have consequently been overlooked by molecular-diversity surveys. In the age of high-throughput analyses, investigation of eukaryotic diversity through culture remains indispensable for the discovery of rare but ecologically and evolutionarily important eukaryotes.
Tuesday, December 6, 2022
Did Archaic Hominins Use Fire?
Ancient DNA From 2000 BCE Hair Of A Herder In Sudan
A new study has analyzed ancient DNA and other chemical features of hair from an individual who died around 2000 BCE and was interred in Sudan. The open access source paper is here. As Bernard explains at this blog (at the link):
This individual is dated 4033 years in the Kerma period which succeeds the Neolithic.A recent study on the diet of individuals buried in Kadruka cemeteries showed that these individuals ate dairy products from cows or sheep, C3-type plants such as beans, cowpeas, cassava, soybeans, rice or barley, or animals eating type C3 plants. The early proliferation of the pastoral economy visible in the Kerma culture of northern Sudan has been proposed as a potential source for the spread of pastoralism in East Africa. . . .
The results show that the ancient individual from northern Sudan is located close to the ancient pastors of East Africa, particularly Kenya and Tanzania, also dated 4000 years ago. Moreover, the f3 statistic shows that this individual from Sudan has the most genetic affinity with the ancient individuals of the Levant, the ancient individuals of North or East Africa, or the current populations of North Africa or the Horn of Africa.These results suggest that the pastoral economy has spread in Africa along the banks of the Nile, towards the south.
Monday, December 5, 2022
Ancient DNA From Sri Lanka
The samples are only mtDNA and are from roughly 3500 BCE and 7500 BCE respectively, both predating the South Indian Neolithic Revolution. Both are mtDNA haplogroups generally associated with higher proportions of Ancient Ancestral South Indian autosomal DNA in modern populations and less old ancient DNA samples.
As expected, continuity from pre-farming times is greater for mtDNA which passes from mother to child, than for Y-DNA for which modern South Asia shows dramatic change from pre-farming eras, since in South Asia, genetic change was associated with conquest by male dominated groups that took local wives more often than colonization by gender balanced families.
Sri Lanka is an island in the Indian Ocean connected by the sea routes of the Western and Eastern worlds. Although settlements of anatomically modern humans date back to 48,000 years, to date there is no genetic information on pre-historic individuals in Sri Lanka.
We report here the first complete mitochondrial sequences for Mesolithic hunter-gatherers from two cave sites. The mitochondrial haplogroups of pre-historic individuals were M18a and M35a. Pre-historic mitochondrial lineage M18a was found at a low prevalence among Sinhalese, Sri Lankan Tamils, and Sri Lankan Indian Tamil in the Sri Lankan population, whereas M35a lineage was observed across all Sri Lankan populations with a comparatively higher frequency among the Sinhalese. Both haplogroups are Indian derived and observed in the South Asian region and rarely outside the region.
A. S. Fernando, et al., "The mitochondrial genomes of two Pre-historic Hunter Gatherers in Sri Lanka" J Hum Genet (November 30, 2022). https://doi.org/10.1038/s10038-022-01099-w
More Ties Between Ordinary And Inferred Dark Matter
Starting from the 1970s, some relations connecting dark matter and baryons were discovered, such as the Tully-Fisher relation. However, many of the relations found in galaxies are quite different from that found in galaxy clusters. Here, we report two new mysterious universal relations connecting dark matter and baryons in both galaxies and galaxy clusters.
The first relation indicates that the total dynamical mass of a galaxy or a galaxy cluster M(500) has a power-law relation with its total baryonic mass M(b) within the `virial region': M(500)∝M(b)^a, with a≈3/4.
The second relation indicates that the enclosed dynamical mass M(d) is almost directly proportional to the baryonic mass for galaxies and galaxy clusters within the central baryonic region: M(d)∝M(b).

The close relations between dark matter and baryons in both galaxies and galaxy clusters suggest that some unknown interaction or interplay except gravity might exist between dark matter and baryons.
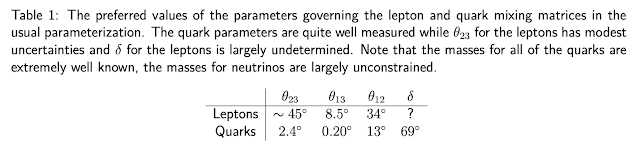
The State Of Neutrino Physics
Scientists are making steady progress in quantifying the properties of the neutrinos. There are seven experiments probing the neutrino oscillation parameters, another seeking to directly measure the absolute value of the lightest neutrino mass, and multiple astronomy collaborations using indirect means to measure neutrino properties including those like IceCube that measure income neutrinos from space directly. As a result of these experiments we are steadily closing the gap of what we know.
The prospects look very good for a precisely known full set of Standard Model neutrino parameters over the next ten to fifteen years, with significant improvements even in the next five years.
In absolute terms, the neutrino masses are already the most precisely known Standard Model parameters and three of the four mixing parameters are also known with decent precision. But, the relative precision with which we know these parameters it the lowest in the Standard Model although this state of affairs may not last too long.
A new new Snowmass 2021 paper has a nice six color chart showing how much progress has been made since 1998 and 2002 when the fact that neutrinos have mass was first discovered.
What we know and don't know is recapped in the executive summary from the Snowmass 2021 paper, the balance of which reviews the various experimental efforts that are underway to answer those questions.
The discovery of neutrino oscillations in 1998 and 2002 added at least seven new parameters to our model of particle physics, and oscillation experiments can probe six of them.
To date, three of those parameters are fairly well measured: the reactor mixing angle θ(13), the solar mixing angle θ(12), and the solar mass splitting ∆m^2(21), although there is only one good measurement of the last parameter. Of the remaining three oscillation parameters, we have some information on two of them: we know the absolute value of the atmospheric mass splitting ∆m^2(31) fairly well, but we do not know its sign, and we know that the atmospheric mixing angle θ(23) is close to maximal ∼ 45º, but we do not know how close, nor on which side of maximal it is. Finally, the sixth parameter is the complex phase δ related to charge-parity (CP) violation, which is largely unconstrained.
Determining these remaining three unknowns, the sign of ∆m^2(31), the octant of θ(23), and the value of the complex phase δ, is of the utmost priority for particle physics. In addition to the absolute neutrino mass scale which can be probed with cosmological data sets, they represent the only known unknown parameters in our picture of particle physics.
It is our job as physicists to determine the parameters of our model. The values of these parameters have important implications in many other areas of particle physics and cosmology, as well as providing insights into the flavor puzzle. To measure these parameters, a mature experimental program is underway with some experiments running now and others under construction. In the current generation we have NOvA, T2K, and Super-Kamiokande (SK) which each have some sensitivity to the three remaining unknowns, but are unlikely to get to the required statistical thresholds. Next generation experiments, notably DUNE and Hyper-Kamiokande (HK) are expected to get to the desired thresholds to answer all three oscillation unknowns. Additional important oscillation results will come from JUNO, IceCube, and KM3NeT. This broad experimental program reflects the fact that there are many inter-connected parameters in the three-flavor oscillation picture that need to be simultaneously disentangled and independently confirmed to ensure that we truly understand these parameters.
To achieve these ambitious goals, DUNE and HK will need to become the most sophisticated neutrino experiments constructed to date. Each requires extremely powerful neutrino beams, as many measurements are statistics limited. Each will require a very sophisticated near detector facility to measure that beam, as well as to constrain neutrino interactions and detector modeling uncertainties, which are notoriously difficult in the energy ranges needed for oscillations. To augment the near detectors, additional measurements and theory work are crucial to understand the interactions properly, see NF06. Finally, large highly sophisticated far detectors are required to be able to reconstruct the events in a large enough volume to accumulate enough statistics. DUNE will use liquid argon time projection chamber (LArTPC) technology most recently demonstrated with MicroBooNE. LArTPCs provide unparalleled event reconstruction capabilities and can be scaled to large enough size to accumulate the necessary statistics. HK will expand upon the success of SK’s large water Cherenkov tank and build a new larger tank using improved photosensor technology.
It is fully expected that with the combination of experiments described above, a clear picture of three-flavor neutrino oscillations should emerge, or, if there is new physics in neutrino oscillations (see NF02 and NF03), that should fall into stark contrast in coming years. . . .
The body text continues to make some useful observations:
The Jarlskog invariant, which usefully quantifies the “amount” of CP violation, for the quark matrix is J(CKM) = +3 × 10^−4 J(max) while for leptons it could be much larger, |J(PMNS)| < 0.34 J(max) where J(max) ≡ 1/(6√3) ≈ 0.096. Understanding this mystery of CP violation is a top priority in particle physics. . . .
There are various other means of probing the six oscillation parameters. In particular, measurements of the absolute mass scale can provide information about the mass ordering in some cases.
• Kinematic end-point experiments such as KATRIN, ECHo, HOLMES, and Project-8 are sensitive to the sum of for all i of |Uei|^2*m(i)^2 which is greater than or equal to 10 meV in the NO and greater than or equal to 50 meV in the IO, although these experiments may not have sensitivity to the mass ordering before oscillation experiments do.
• Cosmological measurements of the cosmic microwave background temperature and polarization information, baryon acoustic oscillations, and local distance ladder measurements lead to an estimate that the sum of for all i of m(i) < 90 meV at 90% CL which mildly disfavors the inverted ordering over the normal ordering since the sum of for all i of m(i) greater than or equal to 60 meV in the NO and greater than or equal to 110 meV in the IO; although these results depend on one’s choice of prior of the absolute neutrino mass scale. Significant improvements are expected to reach the σ(the sum of for all m(ν)) ∼ 0.04 eV level with upcoming data from DESI and VRO, see the CF7 report, which should be sufficient to test the results of local oscillation data in the early universe at high significance, depending on the true values.
• If lepton number is violated via an effective operator related to neutrino mass, then we expect neutrino-less double beta decay to occur proportional to m(ββ) = |the sum of for all i of U(ei)^2*m(i)| which could be as low as zero in the NO but is expected to be > 1 meV in the IO, thus a detection below 1 meV would imply the mass ordering is normal. The latest data from KamLAND-Zen disfavors some fraction of the inverted hierarchy for favorable nuclear matrix element calculations which are fairly uncertain.
• Finally, a measurement of the cosmic neutrino background is sensitive, in principle, to a combination of the absolute mass scale, whether neutrinos are Majorana or Dirac, and the mass ordering.
Among these non-oscillation measurements, only the cosmological sum of the neutrino masses is likely to be sensitive to the atmospheric mass ordering within the next decade.
Consensus is starting to build around (1) a normal mass ordering, (2) a second quadrant (i.e. greater than 45º) value for θ(23), (3) a near minimal value of the lowest absolute neutrino mass, and (4) a complex phase δ for neutrino oscillation CP violation that is non-zero and is close to, but not exactly, maximal.
Neutrinoless double beta decay also remains elusive, if it exists at all. It is undoubtedly already constrained to be very rare, at a minimum and the constraints will continue to grow more strict in the coming years (unless it is finally discovered).
Saturday, December 3, 2022
The Core Theory Physical Constants
The table below has all of the physical constants of the Standard Model of Particle Physics and General Relativity (which are collectively called "Core Theory" in physics) in one place. The data is as of September 2021, although little research since then has been incorporated into the sources cited yet. This version of the table has also been edited from prior versions for greater clarity and stylistic consistency.
Thursday, December 1, 2022
Four New Metric Prefixes Adopted
First uses of prefixes in SI date back to definition of kilogram after the French Revolution at the end of the 18th century. Several more prefixes have gone into use be by the 1947th IUPAC's 14th International Conference of Chemistry, before being officially adopted for the first time in 1960.
The most recent prefixes adopted were ronna-, quetta-, ronto-, and quecto- in 2022, after a proposal from British metrologist Richard J. C. Brown. The large prefixes ronna- and quetta- were adopted in anticipation of needs from data science, and because unofficial prefixes that did not meet SI requirements were already circulating. The small prefixes were added as well even without such a driver in order to maintain symmetry. After these adoptions, all Latin letters have now been used for prefixes or units.
From here.