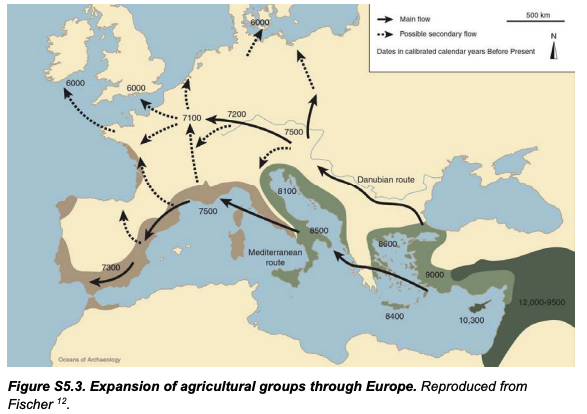
A new preprint analyzes a huge number of new ancient DNA samples from stone age Eurasia (Hat tip to Razib Khan). Despite its claim "Eurasian" scope, it covers pretty much only Europe and part of Siberia. The paper largely confirms and refines existing paradigms and expectations.
LVN = ancestry maximized in Anatolian farmer populations. WHG = ancestry maximized in western European hunter gatherers. EHG = ancestry maximized in eastern European hunter-gatherers. IRN = ancestry maximized in Iranian Neolithic individuals and Caucasus hunter-gatherers.
Razib clarifies beyond the abstract that:
New hunter-gatherer cluster with a focus in the eastern Ukraine/Russian border region. Between the Dnieper and Don. . . .Scandinavia seems to have had several replacements even after the arrival of the early Battle Axe people. This is clear in Y chromosome turnover, from R1a to R1b and finally to mostly I1, the dominant lineage now. They claim that later Viking and Norse ancestry is mostly from the last pulse during the Nordic Bronze Age. . . .it’s clear that Neolithic ancestry in North/Central/Eastern Europe was from Southeast Europe, while that in Western Europe was from Southwest Europe. . . .They find the African R1b around Lake Chad in some Ukrainian samples.
Newly reported samples belonging to haplogroup R1b were distributed between two distinct groups depending on whether they formed part of the major European subclade R1b1a1b (R1b-M269).
Individuals placed outside this subclade were predominantly from Eastern European Mesolithic and Neolithic contexts, and formed part of rare early diverging R1b lineages (Fig. S3b.6). Two Ukrainian individuals belonged to a subclade of R1b1b (R1b-V88) found among present-day Central and North Africans, lending further support5,10 to an ancient Eastern European origin for this clade.
Haplogroup R1b1a1a (R1b-M73) was frequent among Russian Neolithic individuals.
Individuals placed within the R1b-M269 clade on the other hand were from Scandinavian Late Neolithic and early Bronze Age contexts (Fig. S3b.6). Interestingly, more fine-scale sub-haplogroup placements of those individuals revealed that Y chromosome lineages distinguished samples from distinct genetic clusters inferred from autosomal IBD sharing (Fig. S3b.6, S3b.7). In particular, individuals associated with the Scandinavian cluster Scandinavia_4200BP_3200BP were all placed within the sub-haplogroup R1b1a1b1a1a1 (R1b-U106), whereas the two Scandinavian males associated with the Western European cluster Europe_4500BP_2000BP were placed within R1b1a1b1a1a2 (R1b-P312) (Fig. S3b.7).
Figure S3b.6 is as follows and is basically unreadable since even when magnified 500% the resolution isn't fine enough to make it readable, but is presumably referencing the two most basal portions of the circular chart which start in approximately the three o'clock position and evolve counterclockwise. Presumably R1b-V88 is the tree closest to the three o'clock position because it has two samples from the current study in purple/pinkish, while the next tree counterclockwise from it is probably R1b-M73 which has many samples from the study, consistent with the text quoted above.
The footnoted references 5 and 10 to this part of the Supplemental Materials were:
5. Haber, M. et al. "Chad Genetic Diversity Reveals an African History Marked by Multiple Holocene Eurasian Migrations". Am. J. Hum. Genet. 0, (2016). The abstract of this paper states:
Understanding human genetic diversity in Africa is important for interpreting the evolution of all humans, yet vast regions in Africa, such as Chad, remain genetically poorly investigated. Here, we use genotype data from 480 samples from Chad, the Near East, and southern Europe, as well as whole-genome sequencing from 19 of them, to show that many populations today derive their genomes from ancient African-Eurasian admixtures.
We found evidence of early Eurasian backflow to Africa in people speaking the unclassified isolate Laal language in southern Chad and estimate from linkage-disequilibrium decay that this occurred 4,750–7,200 years ago. It brought to Africa a Y chromosome lineage (R1b-V88) whose closest relatives are widespread in present-day Eurasia; we estimate from sequence data that the Chad R1b-V88 Y chromosomes coalesced 5,700–7,300 years ago.
This migration could thus have originated among Near Eastern farmers during the African Humid Period. We also found that the previously documented Eurasian backflow into Africa, which occurred ~3,000 years ago and was thought to be mostly limited to East Africa, had a more westward impact affecting populations in northern Chad, such as the Toubou, who have 20%–30% Eurasian ancestry today.
We observed a decline in heterozygosity in admixed Africans and found that the Eurasian admixture can bias inferences on their coalescent history and confound genetic signals from adaptation and archaic introgression.
and
10. Marcus, J. H. et al. "Genetic history from the Middle Neolithic to present on the Mediterranean island of Sardinia." Nat. Commun. 11, 1–14 (2020) (pertinent Supplemental Information here). The abstract of this paper states:
The island of Sardinia has been of particular interest to geneticists for decades. The current model for Sardinia’s genetic history describes the island as harboring a founder population that was established largely from the Neolithic peoples of southern Europe and remained isolated from later Bronze Age expansions on the mainland.
To evaluate this model, we generate genome-wide ancient DNA data for 70 individuals from 21 Sardinian archaeological sites spanning the Middle Neolithic through the Medieval period. The earliest individuals show a strong affinity to western Mediterranean Neolithic populations, followed by an extended period of genetic continuity on the island through the Nuragic period (second millennium BCE). Beginning with individuals from Phoenician/Punic sites (first millennium BCE), we observe spatially-varying signals of admixture with sources principally from the eastern and northern Mediterranean. Overall, our analysis sheds light on the genetic history of Sardinia, revealing how relationships to mainland populations shifted over time.
The relevant language in the Supplemental Information states:
The 1240k read capture data allowed us to call several R1b subclades in ancient individuals (see Supp. Fig. 7 for overview of available markers). While R1b-M269 was absent from our sample of ancient Sardinians until the Nuragic period, we detected R1b-V88 equivalent markers in 11 out of 30 ancient Sardinian males from the Middle Neolithic to the Nuragic with Y haplogroup calls. Two ancient individuals carried derived markers of the clade R, but we could not identify more refined subclades due to their very low coverage (Supp. Data 1B). The ancient geographic distribution of R1b-V88 haplogroups is particularly concentrated in the Seulo caves sites and the South of the island (Supp. Data 1B).
At present, R1b-V88 is prevalent in central Africans, at low frequency in present-day Sardinians, and extremely rare in the rest of Europe58. By inspecting our reference panel of western Eurasian ancient individuals, we identified R1b-V88 markers in 10 mainland European ancient samples (Fig. 8), all dating to before the Steppe expansion (ą 3k years BCE). Two very basal R1b-V88 (with several markers still in the ancestral state) appear in Serbian HGs as old as 9,000 BCE (Fig. 9), which supports a Mesolithic origin of the R1bV88 clade in or near this broad region. The haplotype appears to have become associated with the Mediterranean Neolithic expansion - as it is absent in early and middle Neolithic central Europe, but found in an individual buried at the Els Trocs site in the Pyrenees (modern Aragon, Spain), dated 5,178-5,066 BCE59 and in eleven ancient Sardinians of our sample. Interestingly, markers of the R1b-V88 subclade R1b-V2197, which is at present day found in Sardinians and most African R1b-V88 carriers, are derived only in the Els Trocs individual and two ancient Sardinian individuals (MA89, 3,370-3,110 BCE, MA110 1,220-1,050 BCE) (Fig. 9). MA110 additionally carries derived markers of the R1b-V2197 subclade R1b-V35, which is at present-day almost exclusively found in Sardinians58.
This configuration suggests that the V88 branch first appeared in eastern Europe, mixed into Early European farmer individuals (after putatively sex-biased admixture60), and then spread with EEF to the western Mediterranean. Individuals carrying an apparently basal V88 haplotype in Mesolithic Balkans and across Neolithic Europe provide evidence against a previously suggested central-west African origin of V88 61. A west Eurasian R1b-V88 origin is further supported by a recent phylogenetic analysis that puts modern Sardinian carrier haplotypes basal to the African R1b-V88 haplotypes 58. The putative coalescence times between the Sardinian and African branches inferred there fall into the Neolithic Subpluvial (“green Sahara”, about 7,000 to 3,000 years BCE). Previous observations of autosomal traces of Holocene admixture with Eurasians for several Chadic populations 62 provide further support for a speculative hypothesis that at least some amounts of EEF ancestry crossed the Sahara southwards. Genetic analysis of Neolithic human remains in the Sahara from the Neolithic Subpluvial would provide key insights into the timing and specific route of R1b-V88 into Africa - and whether this haplogroup was associated with a maritime wave of Cardial Neolithic along Western Mediterranean coasts 63 and subsequent movement across the Sahara 58,64.
Overall, our analysis provides evidence that R1b-V88 traces back to eastern European Mesolithic hunter gatherers and later spread with the Neolithic expansion into Iberia and Sardinia. These results emphasize that the geographic history of a Y-chromosome haplotype can be complex, and modern day spatial distributions need not reflect the initial spread.
The abstract and paper are as follows:
The transitions from foraging to farming and later to pastoralism in Stone Age Eurasia (c. 11-3 thousand years before present, BP) represent some of the most dramatic lifestyle changes in human evolution. We sequenced 317 genomes of primarily Mesolithic and Neolithic individuals from across Eurasia combined with radiocarbon dates, stable isotope data, and pollen records.
Genome imputation and co-analysis with previously published shotgun sequencing data resulted in >1600 complete ancient genome sequences offering fine-grained resolution into the Stone Age populations.
We observe that:
1) Hunter-gatherer groups were more genetically diverse than previously known, and deeply divergent between western and eastern Eurasia.
2) We identify hitherto genetically undescribed hunter-gatherers from the Middle Don region that contributed ancestry to the later Yamnaya steppe pastoralists;
3) The genetic impact of the Neolithic transition was highly distinct, east and west of a boundary zone extending from the Black Sea to the Baltic. Large-scale shifts in genetic ancestry occurred to the west of this "Great Divide", including an almost complete replacement of hunter-gatherers in Denmark, while no substantial ancestry shifts took place during the same period to the east. This difference is also reflected in genetic relatedness within the populations, decreasing substantially in the west but not in the east where it remained high until c. 4,000 BP;
4) The second major genetic transformation around 5,000 BP happened at a much faster pace with Steppe-related ancestry reaching most parts of Europe within 1,000-years. Local Neolithic farmers admixed with incoming pastoralists in eastern, western, and southern Europe whereas Scandinavia experienced another near-complete population replacement. Similar dramatic turnover-patterns are evident in western Siberia;
5) Extensive regional differences in the ancestry components involved in these early events remain visible to this day, even within countries. Neolithic farmer ancestry is highest in southern and eastern England while Steppe-related ancestry is highest in the Celtic populations of Scotland, Wales, and Cornwall (this research has been conducted using the UK Biobank resource);
6) Shifts in diet, lifestyle and environment introduced new selection pressures involving at least 21 genomic regions. Most such variants were not universally selected across populations but were only advantageous in particular ancestral backgrounds. Contrary to previous claims, we find that selection on the FADS regions, associated with fatty acid metabolism, began before the Neolithisation of Europe. Similarly, the lactase persistence allele started increasing in frequency before the expansion of Steppe-related groups into Europe and has continued to increase up to the present. Along the genetic cline separating Mesolithic hunter-gatherers from Neolithic farmers, we find significant correlations with trait associations related to skin disorders, diet and lifestyle and mental health status, suggesting marked phenotypic differences between these groups with very different lifestyles.
This work provides new insights into major transformations in recent human evolution, elucidating the complex interplay between selection and admixture that shaped patterns of genetic variation in modern populations.

1 comment:
Holy Moly! The supplements are 600 pages!! Still in Part 1, Section 2.
Post a Comment